Current Interdisciplinary Research
Sinusoidal Segment Interactive 3D Visualization
Description
Executable file is available upon request.
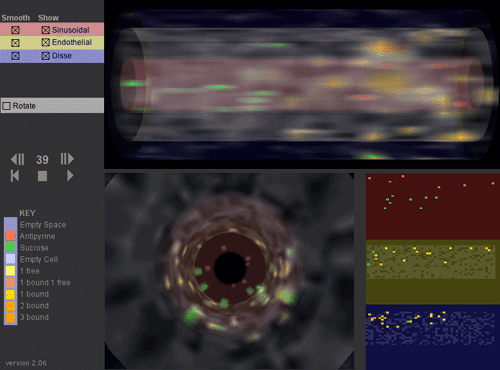
This movie is an interactive 3D visualization of antipyrine & sucrose compounds percolating through one (#7) out of about 60) Sinusoidal Segment in the In Silico Liver (over 400 time steps). The three grid spaces (pink: sinusoidal, yellow: endothelial, and blue: space of the space of Disse plus hepatocytes) are rendered as concentric cylinders. Two views are provided of these cylinders. One is a side view (upper right) and the other is a head-on view (lower left). At the lower right the raw grid data is directly represented.
For each grid space point there are several options:
- It can be empty, that is, no cell there (rendered as fully transparent)
- It can contain an empty cell (rendered as mostly transparent white)
- It can contain antipyrine or sucrose particles outside of a cell (red or green)
- It can contain a cell with one or more free antipyrine particles within it.
- It can contain a cell with one or more bound antipyrine particles within it. For example: "3 bound" means 3 bound antipyrine particles in a cell.
Each "smooth" control on the upper left is used to distinguish the spaces; it causes the grid data to be blurred or not. The blurring is being applied as a texture to a cylinder and could be viewed as representing the likelihood that a compound of that type is actually in that location.
The "Show" control selects the space-sinusoidal, Endothelial and Space of Disse & Hepatocytes (SoD) to be observed (or not).
Selecting "Rotate" causes the cylinder to rotate.
The ">||" "||<" controls causes in silico time to move one step forward or backward.
Notes:
- Compounds are injected into the In Silico Liver using a tight distribution function;
- Antipyrine, but not sucrose, can enter cells;
- There are objects within "cells" that can bind antipyrine;
- Antipyrine bound within "hepatocytes" in the Disse layer can be metabolized.
- Metabolites of antipyrine are not visualized;
- In this movie there is no more than one free solute per cell.
To view the movie you need Macromedia Shockwave Player 10 running on a late-model computer (Mac or Win) with openGL and hardware 3D acceleration.
For additional biological and pharmacokinetic detail see:
Hung DY, Chang P, Weiss M, Roberst MS (2001) Structure-hepatic disposition relationships for cationic drugs in isolated perfused rat livers: transmembrane exchange and cytoplasmic binding process. J Pharmacol Exp Ther. 297:780-9.
Functional Unit Representation Model Project
FURM is a modeling method for building a multitier, in silico apparatus to support iterative experimentation on multiple models.
The development of any given biological model is driven by the experimental context in which it will be used. Hence, computer models are often over-fitted to a single, unique, experimental context and fail to be useful in other situations. To solve this problem, multiple, separate models of a biological system are required to adequately represent that system's behaviors. This is in stark contrast to the biological components to which these models refer, which can be placed in almost any experimental context relevant to their various behaviors. The problem presents a fundamental breakdown in the extent to which any computer model represents its referent. We present the basics of a modeling method, FURM (Functional Unit Representation Method) that attempts to address this breakdown by enforcing and encouraging the use of some basic methodological principles in the development of any functional unit model.
Learn More
Relating GeneChip® Data To Measures of Pharmacological and Toxicological Phenotypes
Can one use microarray measured gene expression (MMGE) data to predict pharmacological and toxicological phenotypes in animals and man? Can similar data be used to account for a significant portion of the individualized variability in patient drug response? In collaboration with the Computational Biology Group at Roche Palo Alto, we are evaluating and contrasting the computational approaches that have been proposed to address these questions. Two large sets of MMGE data are being used to compare and contrast the information that results from use of the various available statistical pattern extraction, clustering and evaluation tools and techniques. Our current focus is on a variable selection and testing strategy that is particularly driven by directly relevant archival phenotypic data, information and knowledge.
Key personnel: Yuanyuan Xiao and Donglei Hu.
Collaborators:
Mark Segal,
Steve Shiboski,
Michael N. Liebman, and Roche Palo Alto.
Toward Genotype-Phenotype Linkage to Optimize Cancer Treatments
This project is part of a larger undertaking that will assemble, test and validate a prototype decision support framework (DSF). We are using both informatics and simulation strategies to create a "mirror" population of hypothetical patients that enables linkage of the information from highly multiplexed molecular analyses through available systems knowledge to patient clinical data so that expected changes in treatment outcomes for new patients can be visualized (on a computer) interactively as various options and/or new data are considered. The DSF is intended to function as part of an institution-wide, patient centered system. With such a system, the patient, the physician, and researchers will be able to detect important, decision impacting relationships contained within the cancer research and clinical trial databases relationships that otherwise might remain undetected. The DSF will also allow the decision makers, the patient and physician, to ask a variety of probing "what if"? questions, and to visually compare and contrast the answers graphically represented, with expected treatment results in real time. Having such a capability will allow customized (and, ideally, optimized) individual treatments, thereby lowering costs while improving both outcomes and the patient's quality of life.
Project contact: C. Anthony Hunt
Collaborators: Oztech Systems, Management Sciences Associates, Dan H. Moore, Joe W. Gray
A Patient-Centered Decision Support Framework for Breast Cancer
This project is a breast cancer specific realization of the project "Toward Genotype Phenotype Linkage to Optimize Cancer Treatments." How can emerging genetic and specific molecular information on breast cancer progression be linked to outcomes associated with different treatments, so that the individual needs of the patient physician interaction can be optimally coordinated? There are critical gaps between the collection of biomedical information (biological and medical informatics) and its most useful application for impacting selection of treatment options. Our goal is to provide an integrated informatics system focused on the key issues in breast cancer treatment that places decision making power in the hands of the clinician, while providing outcome probabilities for the options available to the patient. The DSF (decision support framework) design anticipates making optimum use of such emerging technologies as microarray measures of gene expression. The immediate goal is to demonstrate the feasibility of a DSF that will enable systematic individualization of breast tumor treatments. A computational database is being developed that represents a "mirror" population of breast cancer patients. The core experimental database includes over two dozen prognostic (i.e., breast cancer markers, such as ER status, Her2, etc.), risk, medical, and key patient factors for several hundred patients, plus results from recent pharmacogenetic clinical trials. Each new patient can be matched with a small set "surrogates" within the DSF database, which allows one to ask many "what ifɺ" questions about treatment options and possible outcomes. The next step is to apply and develop algorithms to predict uncertainty throughout the decision making process. In part, our effort is similar to the application of Bayesian Networks to breast cancer. The project anticipates a future clinical setting where breast tumor treatments can be individually optimized, errors will diminish, outcomes will improve, and there will be considerable cost savings for the individual and for society.
Key personnel: Christine Case
Collaborators: John W. Park, Laura J. Esserman, Laurence H. Baker, and Dan Moore.
Using Stochastic Activity Network Simulations to Better Understand Disorders of Hemostasis
Stochastic Activity Networks (SAN) may have advantages over traditional modeling approaches for understanding the complex processes. We are using this tool to better understand the interacting biochemical events involved in the coagulation and fibrinolysis processes that cause Disseminated Intravascular Coagulation! (DIC). We use the UltraSAN2 software environment to simulate these processes. Within the SAN we can inactivate or alter certain proteins to mimic genetic defects and disease, and then compare the results to clinical observations for validation. We are developing this simulation project through 7 steps: 1) trypsinogen activation and trypsin inactivation, 2) normal blood coagulation, 3) disorders of blood coagulation, 4) normal fibrinolysis, 5) disorders of fibrinolysis, 6) normal hemostasis, and 7) DIC. Results of the successful simulations of steps 1?3 by Mounts and Liebman [1]3 provides a promising foundation. A combination of the SAN that separately represents of steps 2 and 4 is expected to adequately represent normal hemostasis. We will systematically inactivate sets of proteins in the final, linked SAN to generate results that are consistent with the clinical observations of DIC. The final, linked SAN will be used to assist in the identification of new therapeutic targets, suggest strategies for development of new diagnostics, and provide a network hypothesis for connecting to emerging genetic information.
Key personnel: Keith Erickson
Collaborators: Michael Liebman and Adam P. Arkin
Mounts, W.M. and Liebman, M.N. Qualitative Modeling of Normal Blood Coagulation and its Pathological States Using Stochastic Activity Networks. Intl J of Biological Macromolecules 20:265-281 (1997).
Elucidating Critical Elements Within Pharmacogenetic and Biomolecular Pathways
We are systematically evaluating the applicability of graph theoretic techniques to facilitate utilization of large pharmacological, genetic and physiological system models. The conceptual or computational model (such the above Stochastic Activity Networks) in is first represented as a network graph having nodes and edges with respective weights. Computational graph analysis is then used to bring critical elements into focus by grouping closely related system elements prior to finding and ranking elements or paths between elements that have the greatest influence on the system. When applied to pharmacological and physiological system models, such "critical elements" represent potential drug targets. By modulating the function of these elements or specific interactions between elements, we expect to be able to identify the process or event where intervention is most likely to have the greatest effect on potential outcome functions, such as drug or treatment effectiveness or undesired side effects.
Key personnel: Abraham Anderson
Collaborators: Richard Ho, Director of Medical Informatics, R.W. Johnson Pharmaceutical Research Institute.
Pharmacogenetics And Individual Variability: Simulation Using Agent-Based Programming.
There is a critical need for new innovative approaches to understanding and modeling interindividual variability and how it impacts development of new therapeutic strategies, e.g., new drugs, as well as the utilization of existing therapeutic strategies. Individual patient variability, such as cardiovascular status, pharmacogenetics, etc. are known to contribute to pharmacodynamic variability. Such variability is difficult to capture using traditional modeling strategies. We are successfully using a novel approach that uses an agent-based programming platform (e.g., SWARM.) The approach can provide an in silico patient population that responds appropriately to drugs based on individual patient attributes. While current approaches in the field rely heavily on complex, often stiff statistical models that are drug specific, our in silico approach offers the ability to model random biological fluctuations inherent in human populations along with a program which may be easily modified to test an array of therapeutic products. The long-term and potential groundbreaking benefits of such an undertaking include targeting appropriate populations for drug safety and efficacy trials, and having a tool that can be used to optimize drug regimens for specific patients.
Key personnel: Amina A. Qutub
Collaborators: Alex Lancaster
A Method for Visualizing Similarity in Gene Expression Datasets
We made significant progress in developing a method of mapping large datasets onto a two dimensional surface where every point corresponds to an Iterated Function Systems (IFS). These IFS can be used to generate an long sequence of scalable arbitrary numbers. There is a correspondence between the similarity of these sequences and their location on the surface of the map. The values contained in these generated sequences is compared against sequential values of target datasets. A point that corresponds to the "best fit" to each target dataset is then located. One can quantify the relationship between similarities of target datasets and the location and geometry of their corresponding points on such a map with a view to optimizing the technique for visualizing relative similarity (or difference) between large sets of gene expression data. Sandy Shaw leveraged the progress made into a new company, Fractal Genomics.
Key personnel: Sandy Shaw
Collaborators: Jenny Harrison (Math UCB).
Microarray Gene Expression In Support of Immunomodulator Discovery
Can one prolong graft survival in transplant patients by using novel therapeutics oligonucleotides to decrease the ability of the graft to trigger the host immune system. Such a strategy is in contrast to the more common approach involving host immunosuppression. We are investigating a series of oligonucleotides that were designed to downregulate the expression of MHC class I and class II genes. In particular, we are interested in the extent to which constitutive and induced expression of MHC genes can be reduced by therapeutic oligonucleotides, and the resultant effect on the host immune response. In Project One, the Hunt Group is implementing strategies to acquire, evaluate, and use microarray measured gene expression to facilitate the above approach.
Collaborators: Richard Shafer
Swarm Modeling and Simulation
Phase 1
Simple Swarm Simulation for Cancer Spheroid
Meeting Minutes
31 May 2002
11:00 a.m. - 1: p.m. HSE-160
Cindy Li and Amy H attending
We began the meeting by discussing a project web page. Cindy agreed to take care of the web page, and we briefly discussed password protecting its contents before deciding against doing so.
We turned our attention to what properties the cells of the tumor should have. Li suggested that we consider oxygen pressure and the area and density of the tumor and create a schedule of objects. Cindy then expressed a desire for a more "top-down" approach, and so we talked about what we wanted the system to do. We agreed that we wanted to simulate growth using a constant growth rate. For the present, we will ignore quiescent and necrotic populations and cell death. I agree it is a good idea to get a simple working program first, even if you know that lots of changes will need to be made. Later, you’ll need to define different cell states, and rules for how they influence each other’s behavior. We also decided that there should be a certain size or number of cells comprising a tumor at which growth ceases, Later you will need to remove this condition. There is no evidence that any one cell is aware of the size (#) of the cell mass that contains it. but we could not reach a consensus on whether size or number of cells should be used. Cells should divide when there is enough nutrient available and at either a certain time or a certain size (we could not reach a consensus on this topic, and it is one that we debated for quite some time). Amy H. also introduced the idea of random cell sizes or random dividing times (e.g., if these conditions are met, then Prob. = p that division will initiate at the start of the step) as having all the cells in the tumor divide at the same time seemed artificial.
At this point we believed that having two classes, a class for cells and a class for the environment, would be the best option, and we attempted to determine rules and properties for the cells and the environment. However, we disagreed about if there should be “levels” for the cells. Cindy thought that the cells and the environment should be on the same level and that their interactions would be mediated by something on a higher level (Is there evidence to support this position?). Amy H. and Li expected that the cells should make “decisions” on their own without such an upper level. Each of us then drew out her own idea of the structure of the program. The concept behind Li’s program was that if there was sufficient amounts of nutrient (N>Nd) and sufficient cell size (S=2S0) but the tumor itself was small enough (it hadn’t reached its limiting size) that a cell would consume some nutrient (Nd) and split into two cells of size S0. Microscopy on spheroids tends to show that daughter cells tend to be smaller after division. They then increase to adult size. Once a daughter cell reaches adult size, there is only a small change in cell size as it then moves through cell the cell cycle. Amy H. questioned the availability of nutrient to a cell at a particular location, and the group agreed to assume that nutrient was freely diffusing through the environment rather than being concentrated at some point.
Cindy still had some concerns about the plan of attack, so we tried to come up with properties and rules that the cells and the environment would have or follow. It became clear that it would be challenging to separate the rules for the environment and the cells. For example when a cell consumes some nutrient, it affects both the cell’s size (it grows) and the amount of nutrient left in the environment. How could we account for both of these consequences? We experimented with the following: if N ≥ Nd, then S=2S0 (the cell would immediately grow to division size) and N=N-Nd. This plan only allows for the splitting of one cell though. Li then suggested that we eliminate the environment class and instead have a fixed amount of nutrient present only as a parameter. However, this did not avoid the problem of how much nutrient is consumed at a particular time step. Important point We realized that we needed to have a count of the cells to determine how much total nutrient is consumed. Our next iteration: at t=0, there are C0 cells and N nutrients. If N ≥ C0Nd, then C0=2C0 (cells split) and N=N-C0Nd (each cell consumes Nd nutrient). If N < Nd, stop. Amy H. pointed out that this strategy left nutrients unused, an unlikely scenario, and Li modified by adding another loop: if Nd ≤ N < C0Nd, C1 = int(N/ Nd ) for some integer C1, so C0= C0+C1 (the C1 cells split). Cindy proposed a different approach where each cell checks the availability of resources (nutrients) for itself only. In other words, if N≥Nd, cell 1 splits. Then if N≥Nd, cell 2 splits, etc. Li thought this approach impractical as cells can split at the same time, but Cindy argued that the tactic was necessary from a programming perspective. Amy H. was displeased with both methods since it seemed unreasonable that a cell would “know” if there was enough nutrient for all cells (as the first approach appeared to suggest) nor did it seem appropriate for each cell to individually wait his turn before splitting.
The meeting concluded with our decision to look into some Swarm examples to get some other ideas for how to proceed and also to determine if it was possible to program things in parallel (so that the cells can do things simultaneously). The group will meet again on Monday at 3:00 p.m. in a location to be announced.
Meeting Minutes
3 June 2002
11:00 a.m. - 1: p.m. HSE-160
Cindy Li and Amy H attending
Li began the meeting by suggesting that we needed to switch tactics given Tony’s comments on Friday. She proposed a single class for the cells with three subclasses (normal, static, and dead) where the subclasses interact with each other. Cindy advised us to start with the normal cells only and add in the static and dead cells later once the program is up and running. When Li and Amy H. asked if that would entail reworking the entire program, Cindy responded that object-oriented programming is designed to allow the addition (and removal) of classes. She then spent some time talking about the heatbug program, and then, having convinced the others that we should begin with the normal subclass of cells only, explained the structure of object-oriented programming using a Swarm tutorial page as a guide.
Cindy asked what properties we wanted the cells to have, and we agreed that the cells should divide if there’s enough room but not if the tumor itself is too large. Amy H. wanted to be sure that the cells would divide in a way such that the tumor remained somewhat spherical (as opposed to dividing down a line), and we discussed the grid structure of Swarm. We thought about considering growth in one-dimension initially and adding the second dimension later, but we believed that such an approach would be difficult to implement.
The idea of using a fixed oxygen gradient was brought up, and we elected to create a class for the environment where we artificially fixed oxygen levels. The goal is for the environment and the tumor to interact with each other in later versions. Cindy recommended that we choose the properties of the cell class and that we split up the work. We determined that the cells should check for available space in four directions (up, down, left, and right, and not in diagonal directions). If space was available, the cell would divide. If more than one space was available, the cell would randomly choose one so that it became unlikely that the tumor would grow in a straight line. The tasks were then divided so that Li and Amy H. would create the classes for the cells and the environment and Cindy would integrate them.
Simulation Modeling of Epithelial Morphogenesis and Malignancy
Epithelial morphogenesis is a fundamental process of development whereby unorganized single cells give rise to coherent forms of tissues, organs and ultimately a whole organism. During morphogenesis, individual cells collectively exhibit self-organizing behaviors as they undergo cycles of growth, migration and differentiation driven by their genetic program and environmental conditions. An essential, yet poorly understood, aspect of morphogenesis concerns with how the various morphogenic processes of single cells are regulated and coordinated to establish recognizable patterns of multicellularity.
Currently we are developing a foundational class of computational models to dissect generative principles of cell action that govern epithelial morphogenesis and malignancy in vitro. The developed models are multi-agent systems composed of autonomous in silico cells whose actions are governed by a set of axioms. Our current study focuses on simulation of two types of epithelial cells: (1) Madine-Darby canine kidney (MDCK) cell growth under varying culture conditions, and (2) primary human alveolar type II (AT II) cell alveolization under 3D culture condition.
Project Contact:Sean H. J. Kim
Collaborators: Wei Yu, Keith E. Mostov, Michael A. Matthay, Jayanta Debnath
Supplementary Material
- In sillico epithellal analogue (ISEA)
Source Codes and supporting files:
- README
- In silico epithelial analogue (ISEA) source codes (download)
- ISEA javadoc(download)
- Compiled ISEA jar files(download)
- A Computational Approach to Understand in Vitro Alveolar Morphogenisis
Source Codes and supporting files:
- README
- AT II analogue source codes (download)
- AT II analogue javadoc (download)
- Compiled AT II analogue jar file (download)
- Simulation Modeling of AT II Cell Wound Healing (in preparation)
Note: simulation space is hexagonal, so when one observes a hexagonal cyst it corresponds to a cross-section through a roundish AT II cyst in vitro. In case of simulated monolayer repair, the in silico visualization corresponds to the xy-planar view in vitro.
- Simulated AT II monolayer wound healing ~12 h [random cell movement]
- Simulated AT II monolayer wound healing ~12 h [chemotaxis]
- Simulated AT II monolayer wound healing ~12 h [cell density-based movement]
- Simulated AT II cyst wound healing [1 wound; 1 cell-width]
- Simulated AT II cyst wound healing [3 wounds; 1 cell-width each]
- Simulated AT II cyst wound healing [1 wound; 3 cell-widths]
- Simulated AT II cyst wound healing [5 wounds; 1 cell-width each]
- Simulated AT II cyst wound healing [1 wound; 5 cell-widths]
- Kim SHJ, Park S, Yu W, Mostov KE, Matthay MA, Hunt CA. 2007. Systems Modeling of Alveolar Morphogenesis In Vitro. ISCA 20th International Conference on Computer and Applications in Industry and Engineering (CAINE 2007), San Francisco, California, USA, November 7-9, 2007

MDCK Cystogenesis Simulation Modeling
Epithelial cells perform essential functions throughout the body, acting as both barrier and transporter and allowing an organism to survive and thrive in varied environments. Although the details of many processes that occur within individual cells are well understood, we still lack a thorough understanding of how cells coordinate their behaviors to create complex tissues. In order to achieve deeper insight, we created a list of targeted attributes and plausible rules for the growth of multicellular cysts formed by Madin-Darby canine kidney (MDCK) cells grown in vitro. We then designed in silico analogues of MDCK cystogenesis using object-oriented programming. In silico components (such as the cells and lumens) and their behaviors directly mapped to in vitro components and mechanisms. We conducted in vitro experiments to generate data that would validate or falsify the in silico analogues and then iteratively refined the analogues to mimic that data. Cells in vitro begin to stabilize at around the fifth day even as cysts continue to expand. The in silico system mirrored that behavior and others, achieving new insights. For example, luminal cell death is not strictly required for cystogenesis, and cell division orientation is very important for normal cyst growth.
Project Contact: Jesse A. Engelberg
Collaborators: Anirban Datta, Keith E. Mostov
In Silico MDCK Analogue Supplementary Material:
- Source code (download)
- User manual